Methods
Imaging30 female and 30 male 8-12-week-old C57BL/6 mice were scanned
in vivo at the Ahmanson-Lovelace Brain Mapping Center at UCLA on a 7T Bruker imaging spectrometer with a micro-imaging gradient insert with a maximum gradient strength of 100 G/cm (Bruker Instruments, Billerica, MA). An actively decoupled quadrature surface coil array was used for signal reception and a 72-mm birdcage coil was used for transmission. Each animal was scanned using a rapid-acquisition with relaxation enhancement (RARE) sequence with the following parameters: TR/TE
eff 3500/32 ms, ETL 16, matrix: 256 x 192 x 100, voxel dimensions: 100 x 100 x 100 µm3. Images were acquired and reconstructed using ParaVision 5.1 software (Bruker Instruments).
Image ProcessingMagnetic resonance images were processed using statistical parametric mapping 8 (SPM8,
http://www.fil.ion.ucl.ac.uk/spm) and the SPMMouse toolbox (
http://www.spmmouse.org) within MATLAB 2013a (Mathworks, Natick, MA). The standard preprocessing steps for voxel-based morphometry (VBM) were carefully adapted as described below to accommodate the analysis of the C57BL/6 mouse brain images acquired
in vivo.
The images were skull-stripped using hand-drawn masks by one rater, bias corrected, and spatially transformed into the same space as the TPMs provided with SPMMouse by applying affine transformations using SPM routines. All images were resliced to match the TPMs with respect to orientation and resolution. The registered and resliced images were segmented using SPMMouse to obtain gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) segments without any linear or non-linear deformations that would encode the shape of the TPMs provided with SPMMouse. These tissue segments were then averaged over all 60 animals and smoothed with a 0.2 mm full-width-half-maximum (FWHM) Gaussian kernel. The new TPMs were then used in a second iteration to align the skull-stripped and bias-corrected images to the reference space and tissue segment these images into GM, WM, and CSF. The resulting segments were averaged and smoothed with a 0.2 mm FWHM Gaussian kernel to generate the final set of what we have named "Mortimer Space" TPMs.
The magnetic resonance images of all 60 mice were manually registered to the newly created TPMs using 6 parameter linear transformations. The images were bias corrected and tissue segmented into GM, WM, and CSF using the unified segmentation algorithm with the newly created TPMs. The resulting tissue segments were used to create a DARTEL template and the individual bias-corrected images were skull-stripped, warped to the DARTEL template, and averaged to create the mean template of the MSA.
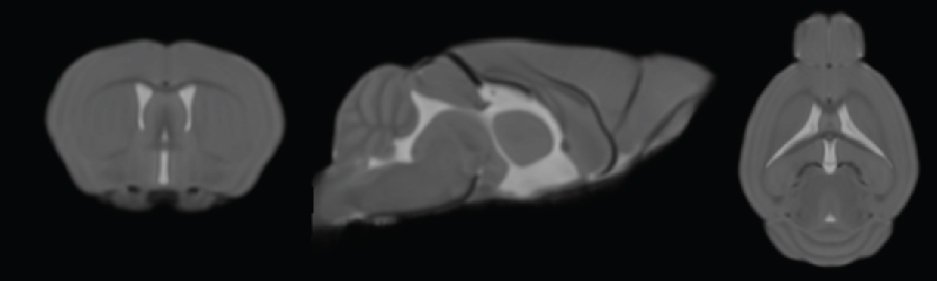
ReferenceMeyer CE, Kurth F, Lepore S, Gao JL, Johnsonbaugh H, Oberoi MR, Sawiak SJ, MacKenzie-Graham A. In vivo magnetic resonance images reveal neuroanatomical sex differences through the application of voxel-based morphometry in C57BL/6 mice. (2017; submitted)


